Part I: Identification and validation of candidate genes on the multi-molecular axis
Differential expression profiles of circRNAs and miRNAs
In the discovery cohort, validation cohort-1, and validation cohort-2, no statistically significant differences in age, sex, and educational years were found (See the detailed statistical results in Supplementary Table 1). High-throughput sequencing discovered 487 DE circRNAs (299 upregulation and 188 downregulation) and 101 DE miRNAs in EOS (16 upregulation and 85 downregulation, Fig. 1A, B). Heatmaps of circRNAs and miRNAs demonstrated that their DE profiles could correctly distinguish between samples from patients with EOS and HCs (Fig. 1C, D). In order to narrow down the list for exploring the actual interactions between genes, and focus on the genes that may have a potentially higher correlation with EOS, we set the values of basic indicators including |fold change | >5 and P < 0.02. The 51 circRNAs were further screened and their predicted binding miRNAs were obtained via the targetscan and miRanda software. By intersecting the predicted list with the DE profile, we discovered that 4 overlapping miRNAs interacted with 10 upstream candidate circRNAs at the prediction level (Fig. 1E).
The volcano maps were drawn to show changes in circRNAs (A) and miRNAs (B) expression profiles. The red dots indicate significant upregulation, blue dots downregulation, and gray dots non-significant genes, with the vertical lines corresponding to fold change and the horizontal line representing P < 0.05. The heatmaps of the DE circRNAs (C) and miRNAs (D) represented their profiles and could correctly distinguish the two kinds of samples. Red indicates higher expression and blue indicates lower expression. E Candidate circRNAs and miRNAs were screened for expressive validation and structural characterization in the next stage of research.
Expressive validation of circRNAs and miRNAs
To verify the reliability of the sequencing data and eliminate the influence of individual heterogeneity, along with the 4 candidate miRNAs and 10 candidate circRNAs selected above, 9 circRNAs and 3 miRNAs were randomly selected from profiles to conduct expressive validation in the validation cohort-1 (40 EOS v.s. 50 healthy controls (HCs)) together. The results showed that 8 out of the 10 candidate circRNAs were statistically different, including chr3:196118684-196129890+,chr1:228743551-228757286-, chrM:2134-2293-, chr21:37711070-37717005-, chrM:1751-1907-, chr12:109046048-109048186+, chr21:37711077-37717005+, and chrM:1689-3208+ (Fig. 2A, B). All candidate miRNAs also showed consistent pathological trends with sequencing data, including hsa-miR-708-3p, hsa-miR-342-3p, hsa-miR-193b-5p, and hsa-miR-20b-5p (Fig. 2C). Simultaneously, 8 out of the 9 randomly selected circRNAs and 3 randomly selected miRNAs have been successfully validated (Supplementary Fig. 1).
A, B The expressive validation results of 10 candidate circRNAs were as follows: chr3:196118684-196129890+ (P = 0.0029), chr1:228743551-228757286-(P < 0.0001), chrM:2134-2293- (P = 0.0054), chr21:37711070-37717005- (P < 0.0001), chrM:1751-1907- (P < 0.0001), chr12:109046048-109048186+ (P = 0.0044), chr21:37711077-37717005+ (P < 0.0001), chrM:1689-3208+ (P < 0.0001), chr1:228743549-228757284-(P = 0.6538) and chr2:148653870-148657467+ (P = 0.0773). C The expressive validation results of 4 candidate miRNAs were as follows: hsa-miR-708-3p, hsa-miR-342-3p, hsa-miR-193b-5p and hsa-miR-20b-5p (P < 0.0001). D Sanger sequencing data showed that head-to-tail of 5 candidate circRNAs matched the expected sizes of circular junctions: hsa-circ-0001189 (TAAT), hsa-circ-0001073 (AAGT), hsa-circ-MORC3 (GTTA), hsa-circ-CORO1C (TTCT) and hsa-circ-TVAS5 (CAAA). E The abundance of chr1:228743549-228757284-wasn’t changed substantially, but the signal was too weak to speculate on enzyme tolerance. Chr21:37711070-37717005- (hsa-circ-MORC3), chr12:109046048-109048186+ (hsa-circ-CORO1C), chr21:37711077-37717005+ (hsa-circ-0001189), chrM:1689-3208+ (hsa-circ-TVAS5) and chr2:148653870-148657467+ (hsa-circ-0001073) were resistant to RNase R (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns not significant, HCs = 50, EOS = 40).
Structural characterization of candidate circRNAs
High stability resulting from the closed-ring structures without 3′ polyA tails and 5′ caps is a crucial property of circRNAs. To confirm whether the candidate circRNAs contain the back-splicing junction region, we amplified the 10 candidate circRNAs via PCR to perform agarose gel electrophoresis (Supplementary Fig. 2a). As shown in the data of Sanger sequencing, the head-to-tail of 5 candidate circRNAs matched the expected sizes of the circular junctions. Among them, except for chr21:37711077-37717005+ (hsa-circ-0001189) and chr2:148653870-148657467+ (hsa-circ-0001073) included in the circbase, none of the other 3 circRNAs have been previously included in databases or studied in EOS. Due to sequence analysis indicating that chr21:37711070-37717005- was formed by back-splicing the MORC Family CW-Type Zinc Finger 3 (MORC3), we named it hsa-circ-MORC3. Similarly, chr12:109046048-109048186+ was called hsa-circ-CORO1C, and chrM:1689-3208+ was named hsa-circ-TVAS5 (Fig. 2D). In addition, RNase R assay revealed the above-mentioned 5 circRNAs were not significantly degraded, indicating that they have higher stability than linear RNAs (Fig. 2E; Supplementary Fig. 2b). The consistent results of Sanger sequencing and RNase R assay confirmed the structural characteristics of these 5 candidate circRNAs.
Part II: Establishment of the multi-molecular axis with binding and regulatory relationships
Binding relationships between circRNAs–miRNAs
Based on validated positive candidate genes and literature review, we further narrowed down the list exploring actual binding relationships to 3 circRNAs and 2 miRNAs. The results showed that co-transfection of hsa-circ-MORC3 and hsa-miR-342-3p or hsa-miR-708-3p did not reduce luciferase activity in HEK293T cells (Fig. 3A, B). Similarly, we did not observe decreased luciferase activity in the co-transfection of hsa-circ-CORO1C or hsa-circ-0001189 with hsa-miR-342-3p (Fig. 3C, D). However, the luciferase activity in the wild-types (WTs) of hsa-circ-CORO1C and hsa-circ-0001189 co-transfected with hsa-miR-708-3p mimic was markedly decreased, while their mutants (muts) were not significantly changed. These suggested that hsa-circ-CORO1C and hsa-circ-0001189, respectively, had stable binding relationships with hsa-miR-708-3p (Fig. 3E, F).
A, B Dual-luciferase reporter assay of HEK293T cells co-transfected with hsa-circ-MORC3 and hsa-miR-342-3p (WT: P = 0.3546, mut: P = 0.1345) or hsa-miR-708-3p (WT: P = 0.0894, mut: P = 0.1000) did not reduce luciferase activity. C, D Co-transfection of hsa-circ-CORO1C (WT: P = 0.1082, mut: P = 0.2000) or hsa-circ-0001189 (WT: P = 0.0674, mut: P = 0.0713) with hsa-miR-342-3p also did not cut down luciferase activity. E, F When hsa-circ-CORO1C (WT: P < 0.0001, mut: P = 0.1012) or hsa-circ-0001189 (WT: P < 0.0001, mut: P = 0.1000) co-transfected with hsa-miR-708-3p, luciferase activity was significantly decreased. G Screening process of target mRNAs predicted to bind with hsa-miR-708-3p. H, I Co-transfection of JARID2 (WT: P = 0.0002, mut: P = 0.1779) or LNPEP (WT: P = 0.0250, mut: P = 0.1955) and hsa-miR-708-3p mimic markedly decreased luciferase activity. J No reduced luciferase activity was observed in the co-transfection of SCAMP1 (P = 0.0594), FAM126B (P = 0.1330), FGR (P = 0.0561), IMPA1 (P = 0.0637), PPP1R3D (P = 0.3045) and hsa-miR-708-3p in HEK293T cells. K After transfected with si-hsa-circ-CORO1C and NC, expression changes of hsa-circ-CORO1C (P < 0.0001), hsa-miR-708-3p (P = 0.0002), JARID2 (P = 0.0127) and LNPEP (P = 0.0042) were detected by qPCR. L After transfected with si-hsa-circ-0001189 and NC, expression changes of si-hsa-circ-0001189 (P = 0.0042), hsa-miR-708-3p (P = 0.4427), JARID2 (P = 0.7476) and LNPEP (P = 0.4838) were detected by qPCR. M, N After transfected mimic of hsa-miR-708-3p into HEK293T cells, hsa-miR-708-3p was upregulated (P = 0.0001) while JARID2 (P = 0.0004) and LNPEP (P = 0.0001) were downregulated. After the transfected inhibitor of hsa-miR-708-3p, hsa-miR-708-3p was downregulated (P = 0.0013) while JARID2 (P = 0.0083) and LNPEP (P < 0.0001) were upregulated.
Binding relationships between miRNAs–mRNAs
Based on the list of possible hsa-miR-708-3p target mRNAs predicted by miRDB and TargetScan, and the list of downregulated mRNAs in EOS, we focused on 7 mRNAs’ functional enrichment analysis significantly related to synapses, negative regulation of gene expression, transcription, phospholipid metabolism, and nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (Fig. 3G). The data showed that co-transfection of JARID2 or LNPEP and hsa-miR-708-3p mimic markedly decreased luciferase activity in HEK293T cells, while the activity did not change when hsa-miR-708-3p binding sites in JARID2 and LNPEP were mutated (Fig. 3H, I). However, co-transfection of SCAMP1, FAM126B, FGR, IMPA1, PPP1R3D, and hsa-miR-708-3p mimic did not reduce luciferase activity in HEK293T cells (Fig. 3J).
Regulatory effects among circRNAs–miRNAs–mRNAs
To further investigate the regulatory effects of circRNAs on miRNA and miRNA on mRNAs, significantly reduced expression of si-hsa-circ-CORO1C indicated successful knockout, while hsa-miR-708-3p was increased and the target JARID2 and LNPEP showed downward trends (Fig. 3K). However, no similar phenomenon was observed when si-hsa-circ-0001189 was transfected into cells (Fig. 3L). In addition, mimic-mediated overexpression of hsa-miR-708-3p significantly inhibited the expression of JARID2 and LNPEP, while JARID2 and LNPEP both increased after being transfected with hsa-miR-708-3p inhibitor (Fig. 3M, N). Through the exploration of the binding and regulatory relationships between candidate circRNAs–miRNAs–mRNAs, we discovered a specific molecular axis for EOS, namely that hsa-circ-CORO1C could function through sponging hsa-miR-708-3p to indirectly regulate the target JARID2 and LNPEP.
Part III: Function of core genes on the multi-molecular axis
Effect of hsa-miR-708-3p on early neurodevelopment
Zebrafish have become an excellent model organism for studying vertebrate genetic function due to their nearly transparent embryos, and 70% homology with the human genome11. Using the characteristic advantages of zebrafish to study the impact of hsa-miR-708-3p would be helpful in understanding its specific role in EOS. Hb9: EGFP zebrafish embryos were divided into the UIC, NC, and overexpression of hsa-miR-708-3p groups after injection (See overall morphology of these three groups under the bright and fluorescent field in Supplementary Fig. 3). At 2-dpf, overexpression of hsa-miR-708-3p led to significant craniocerebral malformation, diminished total brain volume, and smaller orbital development (Fig. 4A). Quantitative analysis of total brain volume also supported these results (Fig. 4B). Images of trunk regions in the hsa-miR-708-3p injection group revealed impaired motor neuron axon growth (Fig. 4C). Quantitative analysis of the average length of spinal motor neuron axons also showed a significant decrease (Fig. 4D).
A Compared to UIC and NC, overexpression of hsa-miR-708-3p led to significant craniocerebral malformation (blue arrowhead) and reduced total brain volume (red area), and smaller orbital development (black arrowhead) at 2-dpf. B The total brain volume of hsa-miR-708-3p was more significantly reduced than that of UIC (P = 0.0003) and NC (P = 0.0006) controls. C Row 1 is the overall morphological appearance of three groups at 2-dpf. Row 2 is imaging of spinal motor neuron structures visualized through fluorescence in the trunk region. Row 3 is magnified views of the boxed regions in row 2, which clearly showed impaired axonal growth in the hsa-miR-708-3p group. D The average length of neuronal axons in hsa-miR-708-3p group was more significantly shortened than that of UIC (P = 0.0002) and NC (P = 0.0009) controls. E At 5-dpf, the percentage of embryos with developmental defects and deaths in hsa-miR-708-3p group was significantly higher than that of UIC (P < 0.0001) and NC (P < 0.0001) controls. F The time-course graph of survival rate within 5 days in three groups. G, H The digital tracks and heatmap image of free exploration aquarium movement tracking experiment at 5-dpf. (I–L) Except for the mobility (UIC v.s. hsa-miR-708-3p, P = 0.2524; NC v.s. hsa-miR-708-3p, P = 0.2664), the total movement distance (UIC v.s. hsa-miR-708-3p, P = 0.0026; NC v.s. hsa-miR-708-3p, P < 0.0001), velocity (UIC v.s. hsa-miR-708-3p, P = 0.0005; NC v.s. hsa-miR-708-3p, P = 0.0005) and acceleration maximum (UIC v.s. hsa-miR-708-3p, P < 0.0001; NC v.s. hsa-miR-708-3p, P = 0.0041) in hsa-miR-708-3p group were significantly lower than controls.
At 5-dpf, the hsa-miR-708-3p injection group had a significantly higher proportion of developmental defects and deaths, and a lower survival rate than these indicators in the UIC and NC controls (Fig. 4E, F). In the free exploration aquarium movement tracking experiment, one larva was placed per well, and each group had ten larva (Fig. 4G, H). The data indicated that except for mobility, locomotor capacities including the total movement distance, velocity, and acceleration maximum in the hsa-miR-708-3p group were significantly lower than those in UIC and NC controls (Fig. 4I–L).
Impacts of JARID2 and LNPEP on neuronal differentiation
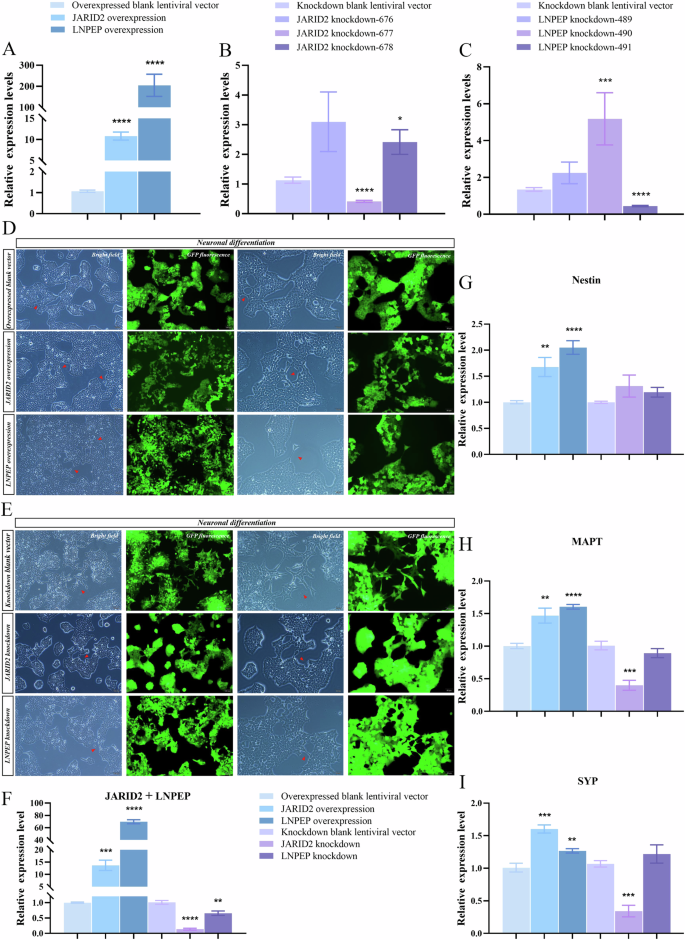
Data analyses showed that both overexpression groups showed significant upregulation (Fig. 5A), knockdown cell line-677 showed significant downregulation of JARID2, and knockdown cell line-491 showed significant downregulation of LNPEP, indicating successful construction (Fig. 5B, C). When the standard medium was just replaced with the neural differentiation medium, cell morphology simply resembled the SH-SY5Y, adhering to the wall in an epithelial-like manner. After 24 h of neural differentiation, neuron-like morphological phenotypes began to appear, with cell bodies retracting to the slightly round shape and an increased number of protrusions. After 96 h, significant morphological changes could be clearly observed. The cell bodies became round or polygonal and a noticeably decreased number of spindle-shaped cells. Partial lengths of the protrusions were longer than the diameter of the cell bodies, forming axon-like and dendrite-like nerve fibers that connected with adjacent cells, building local network-like structures (Fig. 5D, E).
A–C Compared to the blank control, the expression of JARID2 (P < 0.0001) and LNPEP (P < 0.0001) were significantly upregulated in stably transduced overexpression cell lines. The expression of JARID2 was significantly downregulated in cell line-677 (P < 0.0001) and LNPEP was significantly downregulated in cell line-491 (P < 0.0001). D, E Morphological changes during neural differentiation of blank controls, JARID2 and LNPEP stably transduced overexpression and knockdown cell lines (Column 1: 10× bright field, column 2: 10× corresponding fluorescent field, column 3: 20× bright field, column 4: 20× corresponding fluorescent field. Red arrows indicated sites where cell connections formed). F After neural differentiation, JARID2 (P = 0.0001) and LNPEP (P < 0.0001) were significantly upregulated in overexpressed modified cells, while JARID2 (P < 0.0001) and LNPEP (P = 0.0039) were significantly downregulated in knockdown modified cells. G After neural differentiation, nestin was significantly upregulated in JARID2 (P = 0.0064) and LNPEP (P < 0.0001) stably transduced overexpression cell lines, while nestin showed no significant difference in JARID2 (P = 0.1412) and LNPEP (P = 0.0957) stably transduced knockdown cell lines. H, I After neural differentiation, MAPT was significantly upregulated in JARID2 (P = 0.0050) and LNPEP (P < 0.0001) stably transduced overexpression cell lines, while SYP was also significantly upregulated in JARID2 (P = 0.0004) and LNPEP (P = 0.0090) stably transduced overexpression cell lines. MAPT (P = 0.0005) and SYP (P = 0.0004) were significantly downregulated in JARID2 stably transduced knockdown cell line, while MAPT (P = 0.2596) and SYP (P = 0.3026) showed no significant difference in LNPEP stably transduced knockdown cell line.
Morphological change resembling neurons was not sufficient, and biochemical evidence supporting genuine neural responses was also required to determine if JARID2 and LNPEP could significantly affect neuronal development. Due to the circRNAs are highly expressed in the brain and play a crucial role in regulating genes encoding neuronal factors, we used nestin, microtubule-associated protein tau (MAPT), and synaptophysin (SYP) as indicators for evaluating neuronal differentiation outcomes. Firstly, the overexpression and knockdown effects of JARID2 and LNPEP were consistently present during neural differentiation with statistical differences, indicating that significant differences in outcome indicators among the six groups could be caused by JARID2 and LNPEP (Fig. 5F). Nestin showed significant upregulation in both gene overexpression groups, while there was no significant difference in the knockdown groups. These suggested that overexpression of JARID2 and LNPEP could promote the differentiation of SH-SY5Y cells towards the neuronal direction (Fig. 5G). In addition, MAPT and SYP had significant upregulation in both gene overexpression groups, while there was no significant difference in the LNPEP knockdown group. Both MAPT and SYP showed significant downregulation in the JARID2 knockdown group. These results indicated that overexpression of JARID2 and LNPEP could promote axonal growth, increase synaptic density, and contribute to the transmission and processing of neural information (Fig. 5H, I).
Part IV: Clinical roles of the core multi-molecular regulatory axis
For the entire axis, hsa-circ-CORO1C–hsa-miR-708-3p–JARID2 + LNPEP, pathological expression detection, clinical phenotype correlation, and diagnostic efficacy analyses were conducted in validation cohort-2 (84 EOS v.s. 67 HCs). The data indicated that hsa-circ-CORO1C, JARID2, and LNPEP were downregulated in EOS, while hsa-miR-708-3p showed the opposite trend of upregulation with statistical significance (P < 0.05, Fig. 6A). This confirmed again the regulatory relationships of the axis explored through cell transfection experiments. Additionally, the data showed the significant difference in the associations of JARID2 with negative symptoms (P = 0.010), general pathological symptoms (P = 0.008), and total score (P = 0.009), and LNPEP with positive symptoms (P = 0.012), negative symptoms (P = 0.008), general pathological symptoms (P = 0.049), and total score (P = 0.012, Fig. 6B, C).
A Hsa-circ-CORO1C (P = 0.0022), JARID2 (P < 0.0001) and LNPEP (P = 0.0415) were significantly downregulated, while hsa-miR-708-3p (P = 0.0254) was significantly upregulated (HCs = 67, EOS = 84; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). B, C JARID2 was significantly associated with negative symptoms (r = −0.312, P = 0.010), general pathological symptoms (r = −0.322, P = 0.008), and total score (r = −0.317, P=0.009), while LNPEP was significantly associated with positive symptoms (r = −0.283, P = 0.012), negative symptoms (r = −0.298, P = 0.008), general pathological symptoms (r = −0.224, P = 0.049) and total score (r = −0.284, P = 0.012). D, E Diagnostic efficacy evaluation of hsa-circ-CORO1C (AUC = 0.6563, 95% CI: 0.5440~0.7685, P = 0.0023) and hsa-miR-708-3p (AUC = 0.6031, 95% CI: 0.5107~0.6954, P = 0.0257). F–H Diagnostic efficacy evaluation of JARID2 (AUC = 0.7056, 95% CI: 0.6077~0.8035, P < 0.0001), LNPEP (AUC = 0.5989, 95% CI: 0.5034~0.6944, P = 0.0415) and the combined diagnostic efficacy of JARID2 and LNPEP (AUC = 0.7727, 95% CI: 0.6815~0.8638, P < 0.0001). I Combined diagnostic efficacy evaluation of the whole multi-molecular axis (AUC = 0.8037, 95% CI: 0.7030~0.9044, P < 0.0001). J Schematic illustration of diminished total brain volume, restricted axonal growth, and impacted synaptic density caused by hsa-circ-CORO1C–hsa-miR-708-3p–JARID2 + LNPEP axis with great diagnostic and evaluative abilities in EOS.
The results of diagnostic efficacy evaluation revealed that hsa-circ-CORO1C (AUC = 0.6563, sensitivity = 90.54%, specificity = 69.64%) and hsa-miR-708-3p (AUC = 0.6031, sensitivity = 88.42%, specificity = 47.76%) possessed relatively good diagnostic power (Fig. 6D, E). JARID2 (AUC = 0.7056, sensitivity = 78.87%, specificity = 71.43%) and LNPEP (AUC = 0.5989, sensitivity = 50%, specificity = 79.37%) had a combined diagnostic efficacy (AUC = 0.7727, sensitivity = 80.28%, specificity = 78.95%) superior to the individual mRNAs (Fig. 6F–H). Furthermore, the combined diagnostic efficacy of the whole multi-molecular axis (AUC = 0.8037, sensitivity = 91.55%, specificity = 84.78%) was higher than the combined diagnostic efficacy of JARID2 and LNPEP (Fig. 6I).
Combined evidence from genetic, neurophenotypic, and clinical perspectives, at the pathological microscale, EOS exhibited a dysregulation pathway in hsa-circ-CORO1C acted as a sponge to attenuate hsa-miR-708-3p’s inhibitory effects on target JARID2 and LNPEP. At the structural mesoscale, this aberrant multi-molecular regulatory axis caused neurodevelopmental phenotypes of diminished total brain volume, restricted axonal growth, and decreased synaptic density. At the functional macroscale, multiple genes on the regulatory axis demonstrated great recognition ability in the multidimensional clinical symptoms of EOS. The combined diagnostic efficacy of the entire axis was superior to that of any individual gene we discovered in EOS (Fig. 6J).